Hi there! Welcome to my webpage.

I am a researcher with interdisciplinary aptitude having chemical engineering background followed by strong research accomplishment in computational chemistry and biophysics. My research interests involve applying the super-computing facilities to understand the relationship between structure and function of biomolecules like proteins, nucleic acids, carbohydrates and small drug-like molecules.
Currently, I am leading multiple projects to develop and apply an advanced structure-based drug design platform called SILCS. I am always interested in integration of computational approach towards understanding the functions of biomolecules and their interactions with drug-like small molecules into experimental methods, eventually achieving faster and better development of clinical candidates.
Research interests
# Molecular Modeling # Protein-ligand interactions # Protein-protein interactions # RNA folding, structures and functions # Small-molecule Drug Discovery # Structure Function Relationships # Free energy calculations # Enhanced sampling # Shell scriptingPublications
-
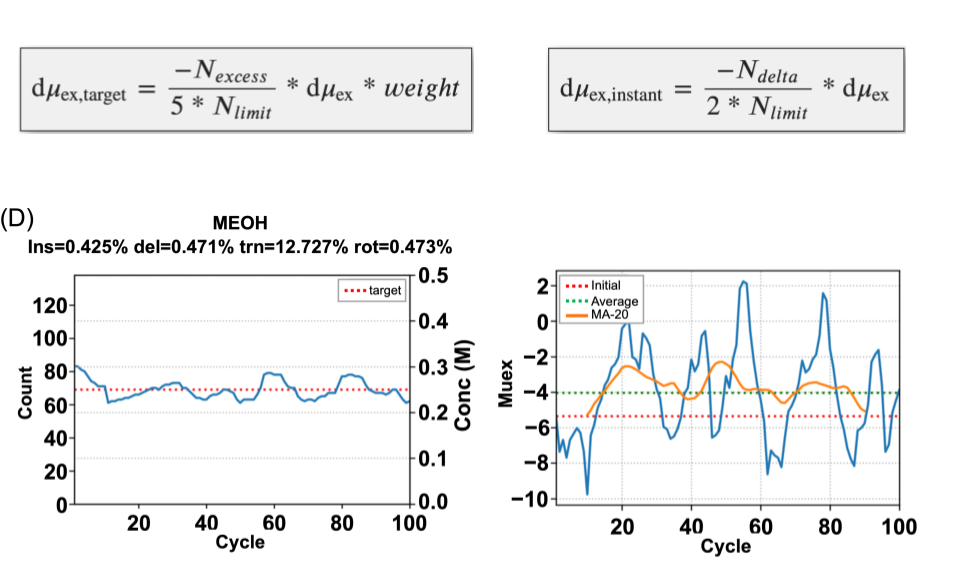
GPU-specific algorithms for improved solute sampling in grand canonical Monte Carlo simulations
Zhao M; Kognole AA ; Jo S ; Tao A ; Hazel A; MacKerell AD Jr.
Journal of Computational Chemistry (First published on Apr 24, 2023)
10.1002/jcc.27121
[PDF]
-
Extension of the CHARMM Classical Drude Polarizable Force Field to N- and O-linked Glycopeptides and Glycoproteins
Kognole AA ; Aytenfisu A ; MacKerell AD Jr.
Journal of Physical Chemistry B (First published on Aug 25, 2022)
10.1021/acs.jpcb.2c04245
[PDF]
-
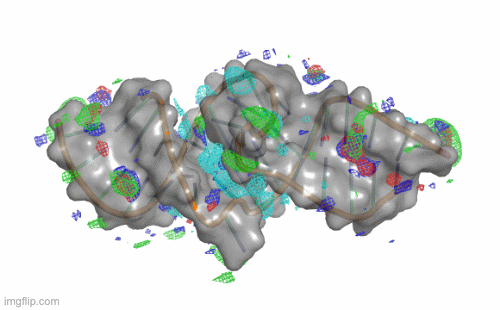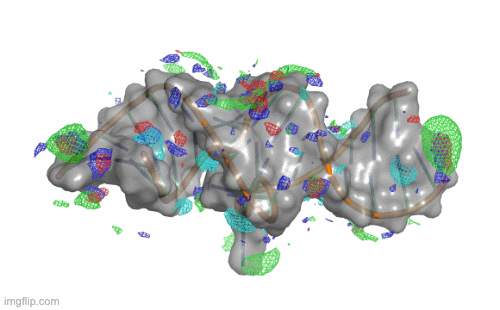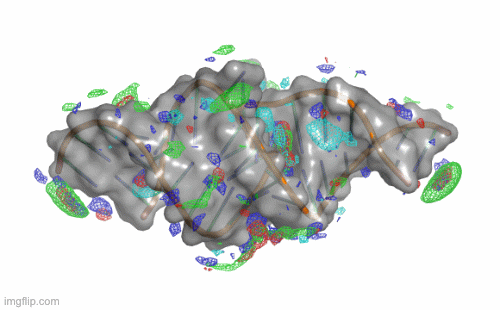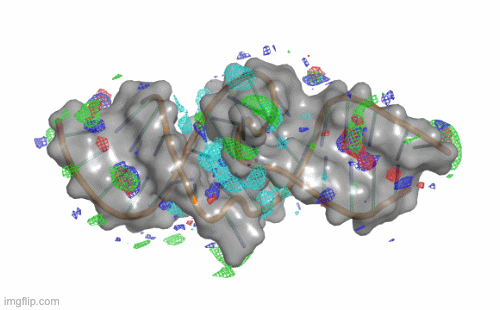
SILCS-RNA: Toward a Structure-Based Drug Design Approach for Targeting RNAs with Small Molecules
Kognole AA ; Hazel A ; MacKerell AD Jr.
Journal of Chemical Theory and Computation (First published on Aug 1, 2022)
10.1021/acs.jctc.2c00381
[PDF]
-
CHARMM-GUI Drude Prepper for Molecular Dynamics Simulation Using the Classical Drude Polarizable Force Field
Kognole AA ; Lee J ; Park SJ ; Jo S ; Chaterjee P ; Lemkul JA ; Huang J ; MacKerell AD Jr. ; Im W
Journal of Computational Chemistry (2022) 43(5):359-375
10.1002/jcc.26795
[PDF]
-
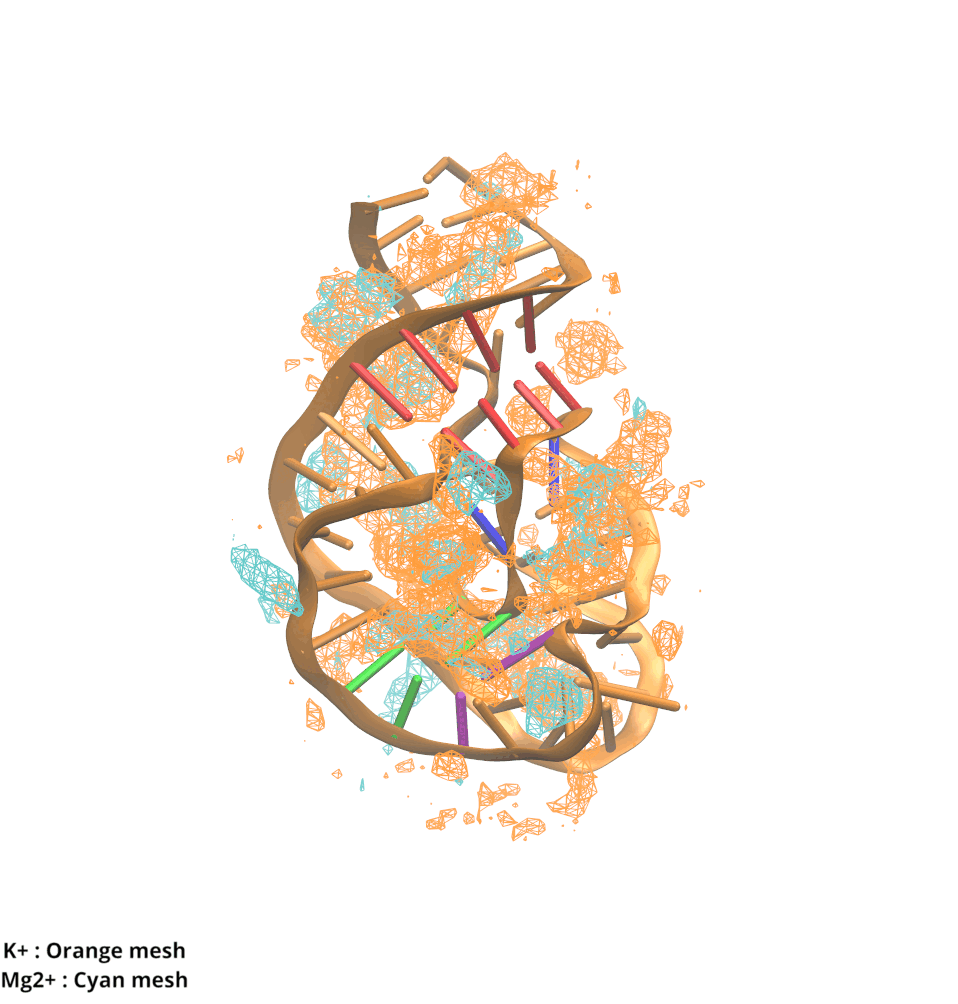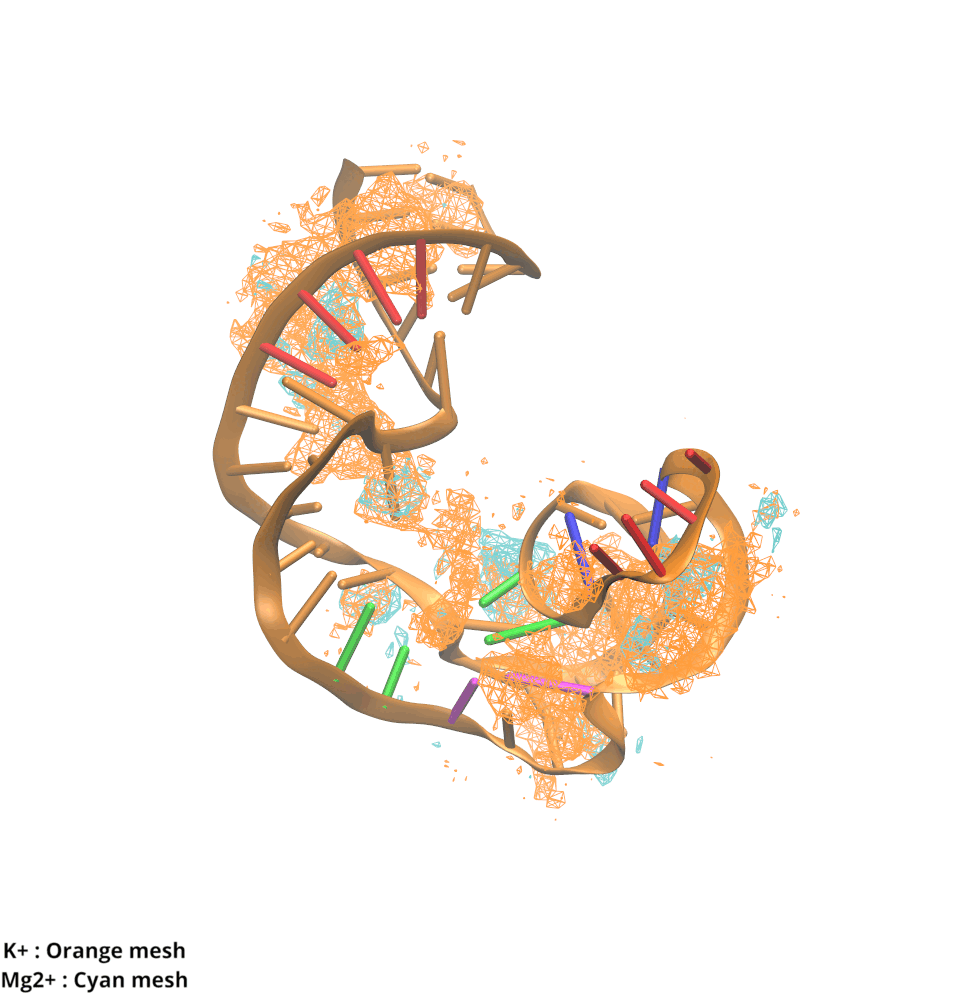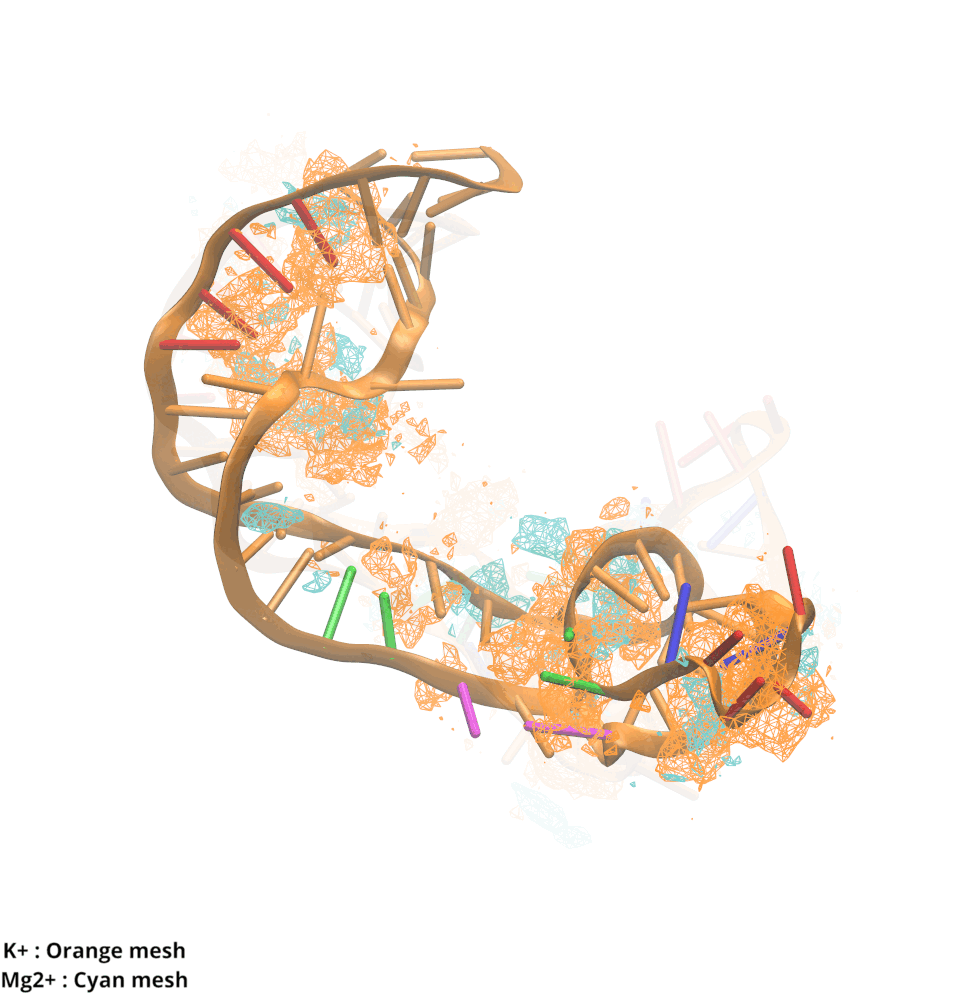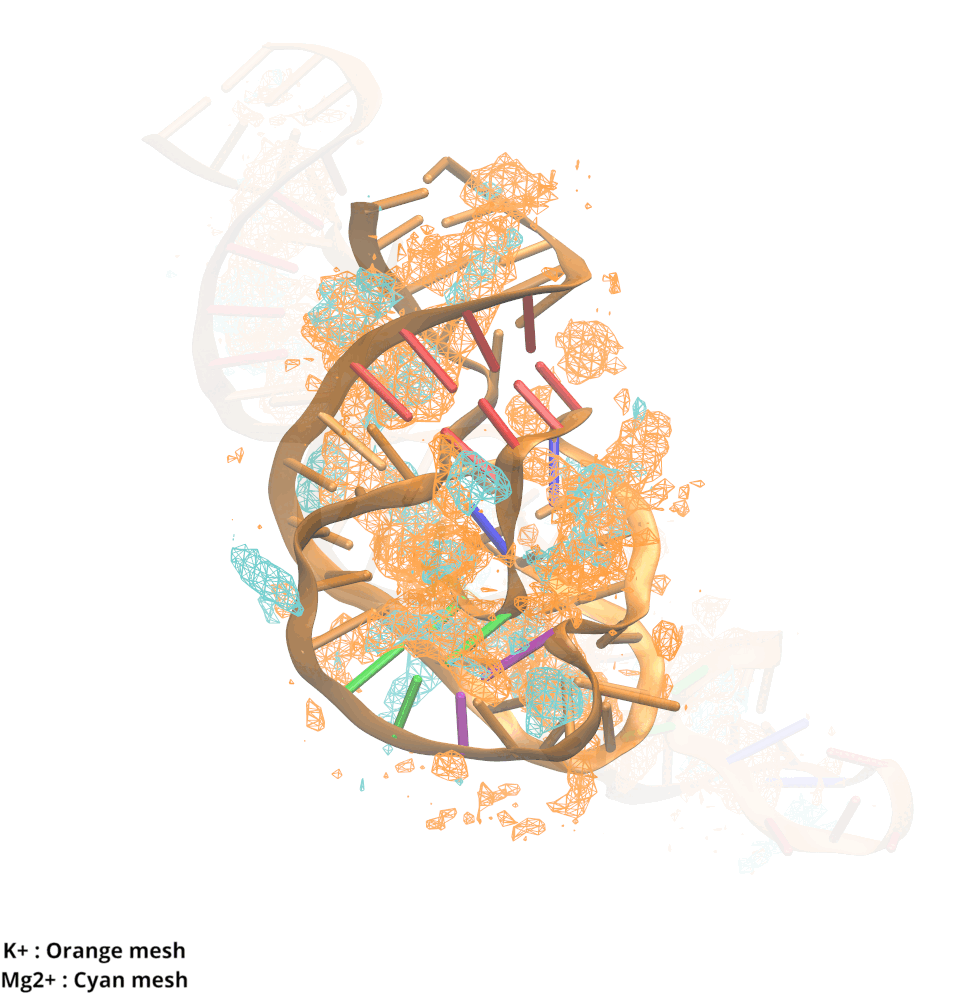
Contributions and Competition of Mg2+ and K+ in Folding and Stabilization of the Twister Ribozyme
Kognole AA ; MacKerell AD Jr.
RNA (2020) 26(11):1704-1715
10.1101/2020.06.15.152744
[PDF]
a href="https://github.com/aakognole/rna-mg-predictor">
-
Balanced Polarizable Drude Force Field Parameters for Molecular Anions: Phosphates, Sulfates, Sulfamates and Oxides
Kognole AA ; Aytenfisu AH ; MacKerell AD Jr.
Journal of Molecular Modeling (2020) 26(152)
10.1007/s00894-020-04399-0
[PDF]
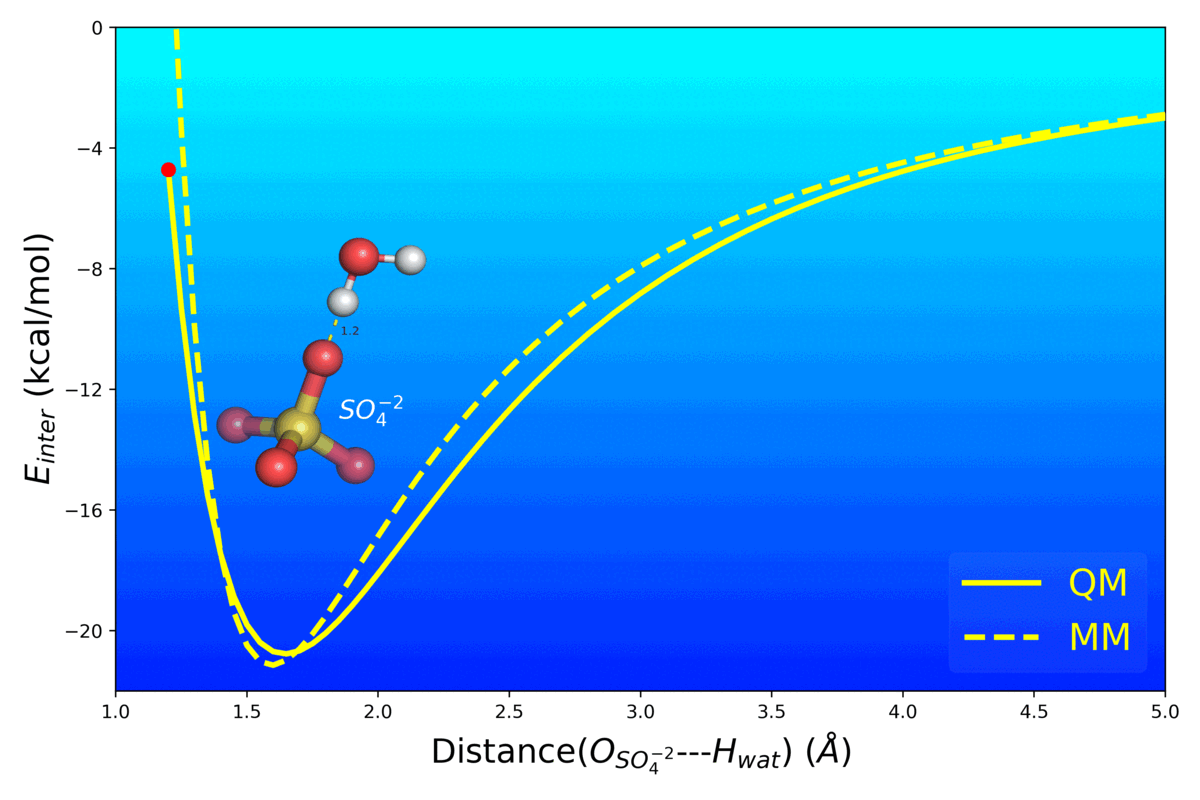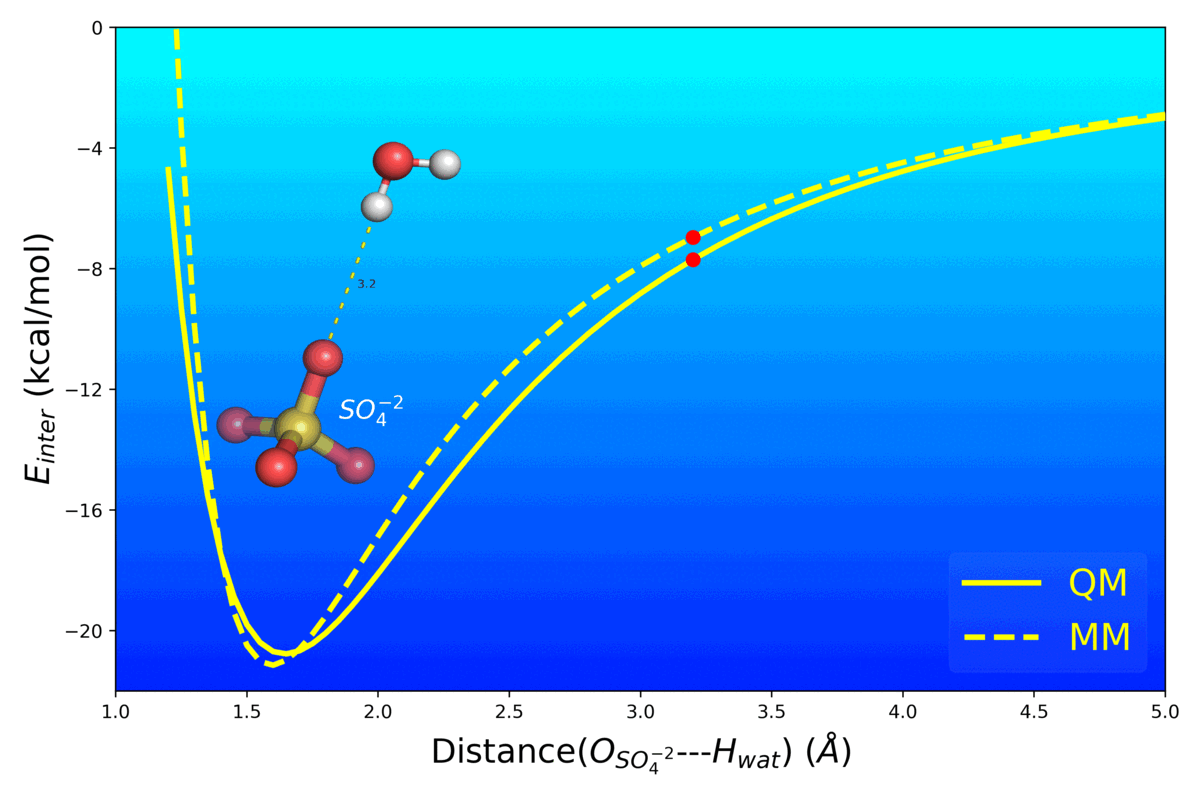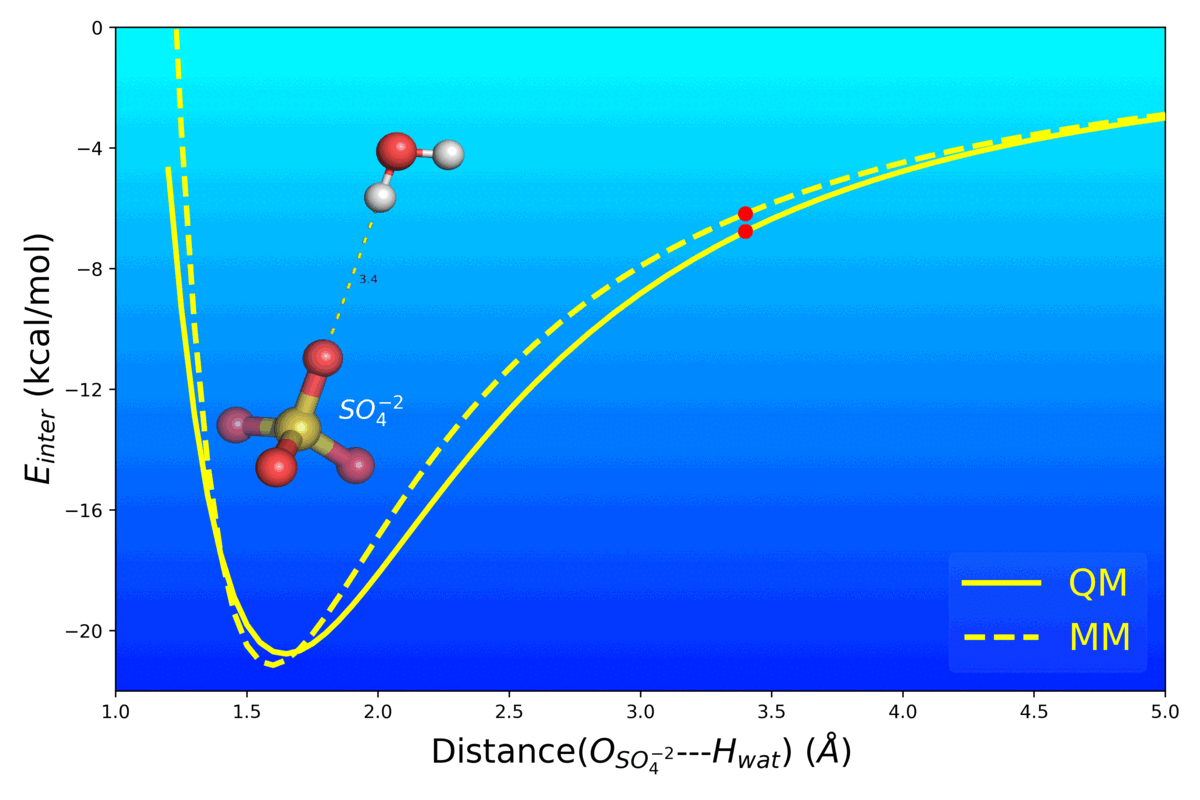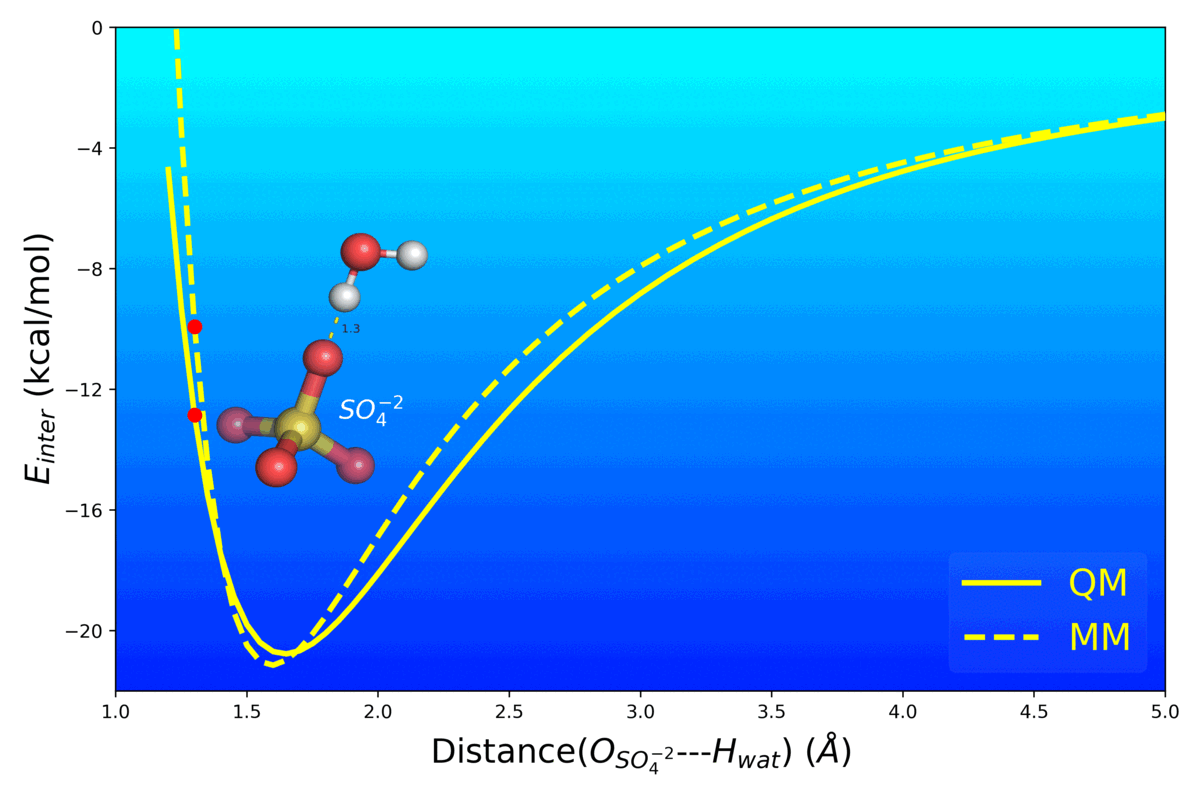
- Mg2+ Impacts the Twister Ribozyme Through Push-Pull Stabilization of Non-Sequential Phosphate Pairs Kognole AA ; MacKerell AD Jr. Biophysical Journal (2020) 118(6): 1424-1437 10.1016/j.bpj.2020.01.021 [PDF]
- Predicting Partition Coefficients of Neutral and Charged Solutes in the Mixed SLES–Fatty Acid Micellar System Turchi M ; Kognole AA ; Kumar A ; Cai Q ; Lian G ; MacKerell AD Jr. Journal of Physical Chemistry B (2020) 124(9): 1653-1664 10.1021/acs.jpcb.9b11199 [PDF]
- Cellulose-specific Type B Carbohydrate Binding Modules: Understanding Substrate Recognition Mechanisms Through Molecular Simulation Kognole AA ; Payne CM Biotechnology for Biofuels (2018) 11(319) 10.1186/s13068-018-1321-7 [PDF]
- Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition Liu B ; Kognole AA ; Wu M ; Westereng B ; Crowley MF ; Kim S ; Dimarogona M ; Payne CM ; Sandgren M FEBS Journal (2018) 285(12): 2225-2242 10.1111/febs.14472 [PDF]
- Inhibition of the Mammalian Glycoprotein YKL-40: Identification of the Physiological Ligand Kognole AA ; Payne CM Journal of Biological Chemistry (2017) 292(7): 2624-2636 10.1074/jbc.M116.764985 [PDF]
- Structural and Functional Characterization of a Lytic Polysaccharide Monooxygenase with Broad Substrate Specificity Borisova AS ; Isaksen T ; Dimarogona M ; Kognole AA ; Mathiesen G ; Várnai A ; Røhr ÅK ; Payne CM ; Sørlie M ; Sandgren M ; Eijsink VGH Journal of Biological Chemistry (2015) 290(38): 22955-22969 10.1074/jbc.M115.660183 [PDF]
- Cello-oligomer-binding dynamics and directionality in family 4 carbohydrate-binding modules Kognole AA ; Payne CM Glycobiology (2015) 25(10): 1100-1111 10.1093/glycob/cwv048 [PDF]
Presentations
19. Rouen KC, Ngo K, Kognole AA, Dawson JRD, Bekker B, Yarov-Yarovoy V, MacKerell AD Jr., Vorobyov I, "Exploring drug arrythmogenicity via atomistic simulations of state-specific hERG-channel block" Biophysical Society Annual Meeting 2022, San Fransisco, CA, USA; Feb 2022. (P)
18. Kognole AA and MacKerell AD Jr., "Development and Evaluation of the Silcs Methodology for Targeting RNA with Small Molecules" Biophysical Society Annual Meeting 2021, Conducted Online; March 2021. (P)
17. Sengul MY, Kognole AA and MacKerell AD Jr., "Enhanced Sampling of RNA Folding/Unfolding Configurations using Machine Learning/Grand Canonical Monte Carlo" Biophysical Society Annual Meeting 2021, Conducted Online; March 2021. (P)
16. Kognole AA and MacKerell AD Jr., "Changes in the ionic atmosphere impact the transition from compact intermediates to the folded state in Twister RNA" American Chemical Society Annual Meeting 2020, Conducted Online; March 2020. (O) (Click here for slides)
15. Kognole AA and MacKerell AD Jr., “The Role of Mg+2 Ion Interactions in Folding of the Twister Ribozyme and PreQ1 Riboswitch Revealed through Umbrella Sampling Combined with Oscillating Chemical Potential Grand Canonical Monte Carlo/Molecular Dynamics Simulations” Biophysical Society Annual Meeting 2019, Baltimore, MD, USA; March 2019. (Poster)
For more click here
...
Work Experience
- Apr 2022 - Present, Applications Scientist, Computational Chemistry, Early Charm Ventures (SilcsBio LLC)
- Nov 2017 - Apr 2022, Postdoctoral Research Fellow, University of Maryland Baltimore (MacKerell lab)
- Aug 2012 - Oct 2017, Graduate Research Assistant, University of Kentucky (Payne lab)
Education
- Aug 2012 - Sept 2017,Ph.D. in Chemical Engineering, Chemical & Materials Engineering, University of Kentucky (Payne Lab) [Thesis]
- Jul 2008 - May 2012, B.E. in Chemical Engineering, Institute of Chemical Technology (Formerly UDCT), Mumbai